The eggplant high quality genome sequence has been made publicly available:
solgenomics.net/organism/Solanum_melongena/genome.
It has been obtained by an Italian consortium including University of Torino (DISAFA, Plant Genetics and Breeding, team leader Sergio Lanteri), CREA (Council for Agricultural Research and Economics, Research Centre for Genomics and Bioinformatics, Montanaso Lombardo, team leader Giuseppe Leonardo Rotino), University of Verona (Functional Genomics Lab, Department of Biotechnology, team leader Massimo Delledonne), and ENEA (Italian National Agency for New Technologies, Energy and Sustainable Development, Casaccia, team leader Giovanni Giuliano). The research was financed by the private seed companies Enza Zaden, Rijk Zwaan and Vilmorin.
- Barchi et al. (2019): A chromosome-anchored eggplant genome sequence reveals key events in Solanaceae evolution. Scientific Reports, (9):11769
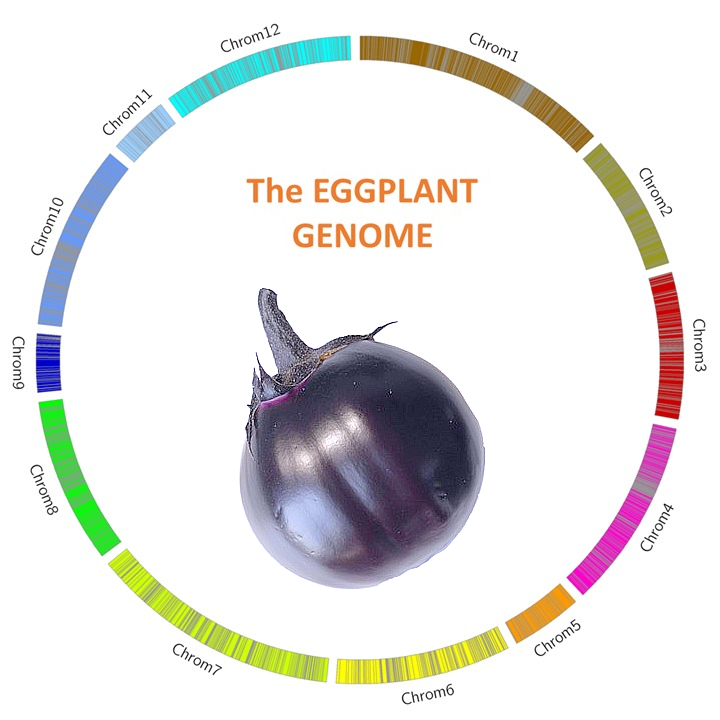
The genomic sequence was obtained by an innovative approach which combines two methodologies. Firstly, from ~257-fold next-generation sequencing (NGS) data, about 23K sequences (N50= 640 Kbp) were assembled. Then, by using the Bionano Genomics optical mapping, an optical map of the reference genome was built up and combined with the NGS data, resulting in the final de novo assembly. It’s size is about 1.18 Gbp, with a map N50 of 2.09 Mb and including 39,922 genes. Thanks to a low coverage whole genome-sequencing of an F6 RIL mapping population and using a novel pipeline, namely SOILoCo (Scaffold Ordering by Imputation with Low Coverage), already applied to the artichoke genome sequencing project (http://www.artichokegenome.unito.it), a dense genetic map based was built up and used for anchoring the hybrid assembly. The final eggplant pseudomolecules span 1.2Gb of the eggplant genome, with a N50 value of 3.22Mb.
Bioinformatics analyses have also provided insights on the eggplant genome and used to conduct comparative analyses among S. melongena and the three other most important Solanaceae species (potato, tomato and pepper), including protein coding genes along with distribution of gene families.
To gain benefit of the whole genome sequence the bulk mining of its markers in totality is imperative and critically required. Here we introduce the first microsatellite marker database EgMiDB (Eggplant MicroSatellite DataBase) based on whole genome SSR mining.

http://dx.doi.org/10.3389/fpls.2018.00401

Conference Abstract

Conference website

Conference Abstract
