
SSR mining
Perfect, imperfect and compound SSRs were in-silico mined using the SciRoKo SSR-search module (http://kofler.or.at/bioinformatics/SciRoKo) within pseudomolecules as well as unmapped scaffolds of the recently developed high quality reference eggplant genome. A minimum of four repetitions together with a minimum length of 15nt was requested; so any sequence was considered as a perfect SSR where a motif was repeated at least 15 times (1nt motif), eight times (2nt), five times (3nt) or four times (4-6nt), allowing for only one mismatch. For compound repeats, the maximum default interruption (spacer) length was set at 100bp.
SSR motif frequency and distribution
From the ~1.1 Gb of the gap-free eggplant genomic sequence, we identified 132,831 perfect SSR motifs (density of about 120 SSR/Mb), which included 20,760 (15.6%) compound SSRs. The imperfect SSR motifs identified were over 178,000 (Table 1). Dinucleotides were the most, representing 42.8% of all microsatellites, followed by tri- (37.0%), mono (8.4%) and tetranucleotides (7.1%). Penta- and hexanucleotide repeats were the least frequent SSR types, together representing less than 5% of the set of perfect SSRs. A/T, AT/AT, AAC/GTT, AAAT/ATTT, AAAAT/ATTTT and AACAAT/ATTGTT were the most frequent repeats among mono- to hexanucleotide SSRs (Table 2).
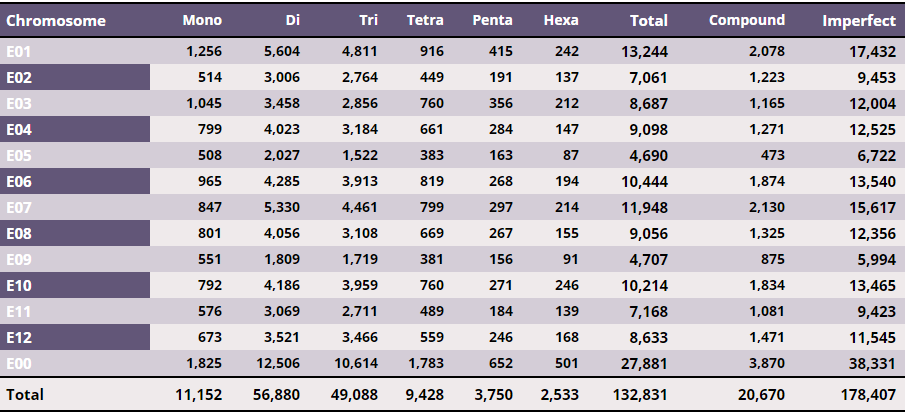
Table 1 – Chromosome wise distribution of perfect SSRs
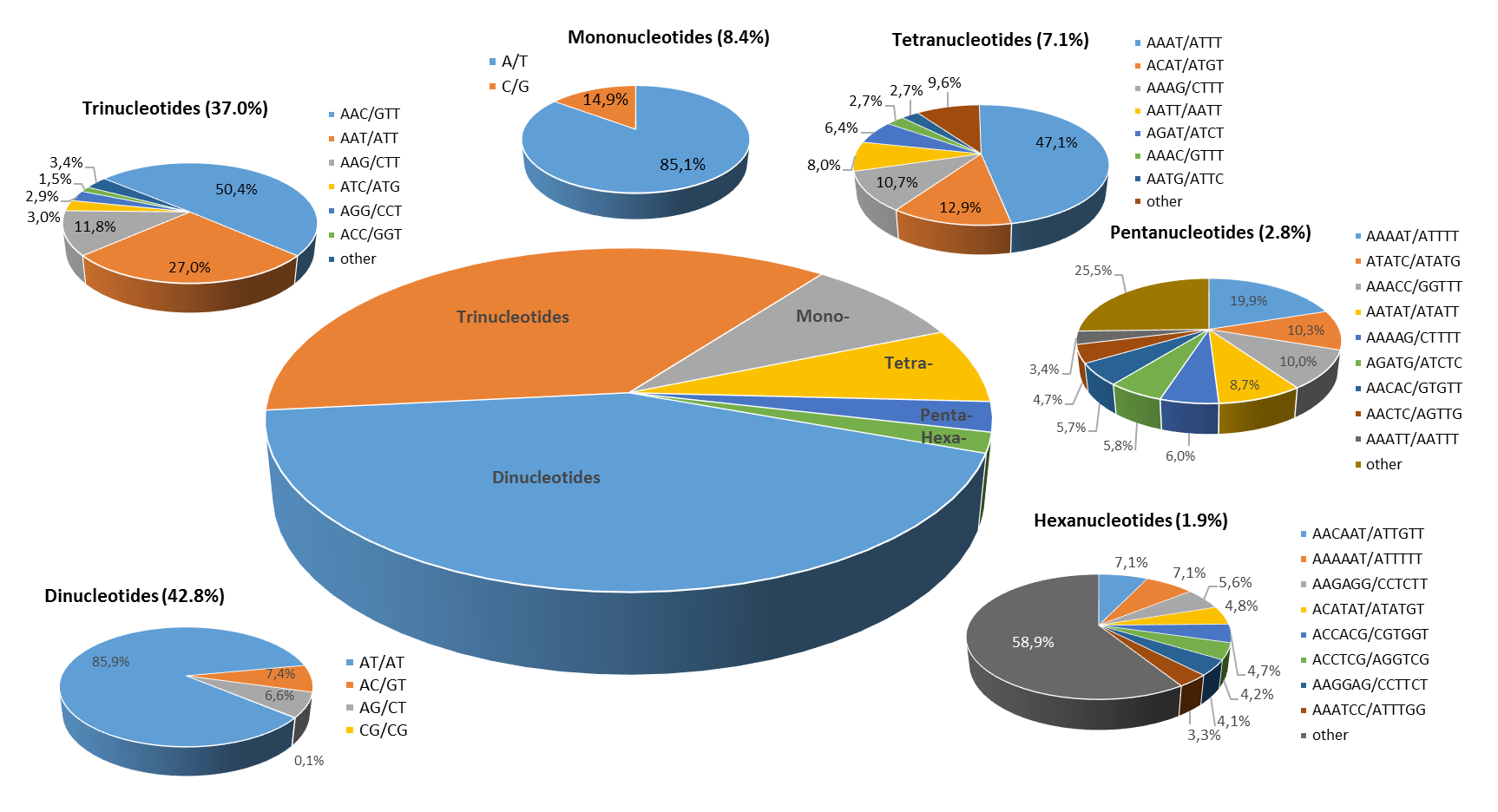
The distribution of the major repeat types in the eggplant genome
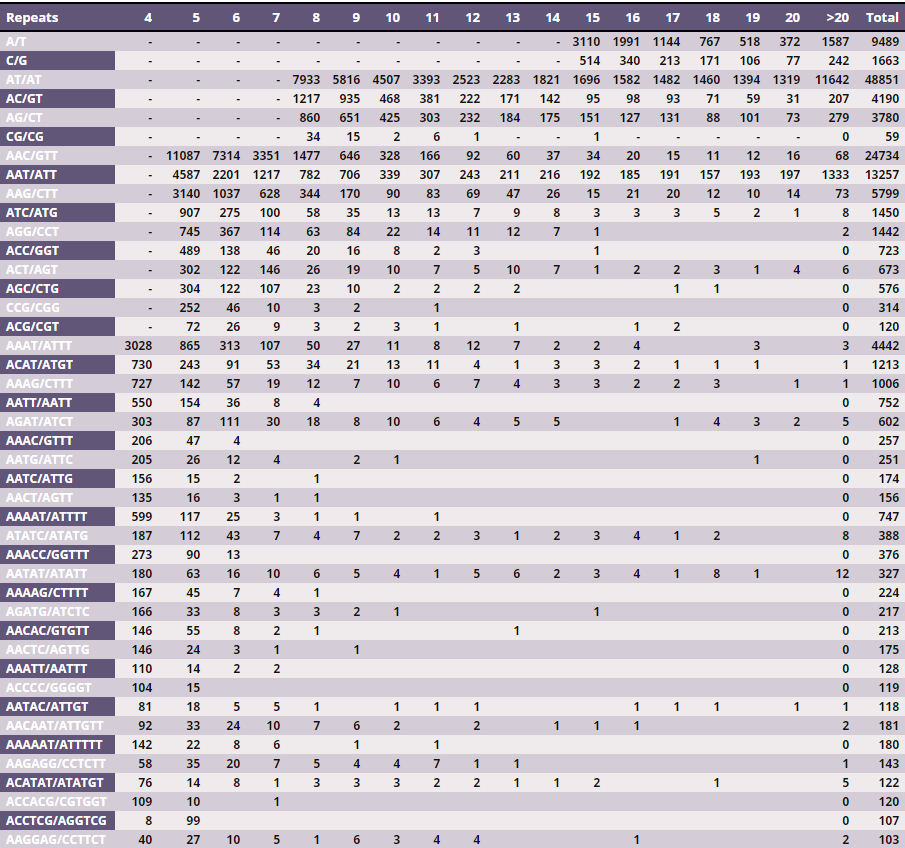
Table 2 – Frequency of the main identified SSR motifs (considering sequence complementary)
